b1-correction
Correction of B1 inhomogeneity artifacts in CEST images
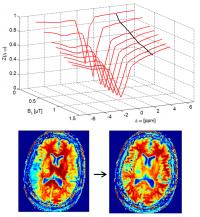 Here you find a B1-correction algorithm as published in Windschuh et al. (2015), NMR Biomed., 28(5):529-37. doi: 10.1002/nbm.3283.
Here you find a B1-correction algorithm as published in Windschuh et al. (2015), NMR Biomed., 28(5):529-37. doi: 10.1002/nbm.3283.
Dowload zipped Matlab implementations here or find the package on https://github.com/cest-sources/B1_correction/
Tutorial
There are two functions, Z_B1_correction and contrast_B1_correction, which both require data of several CEST acquisitions with different B1. Both reconstruct either Z-spectrum stacks or contrast stacks at the B1 values given by B1_output.
function [Z_stack_corr] = Z_B1_correction(Z_stack,rel_B1map,B1_input,B1_output,SEGMENT,fit_type,B1_input_index) % [Z_stack_corr] = Z_B1_correction(Z_stack,rel_B1map,SEGMENT_2D,B1_input,fit_type,B1_input_index,B1_output) % output: B1 corrected Z-stack (5D-stack) % input: Z_stack = 5D-stack of Z-spectra (y,x,z,offset,B1) % rel_B1map = relative B1map % B1_output = output B1 (scalar or vector) % SEGMENT = voxel/pixel mask for evaluation % fit_type = fit or interpolation type % B1_input_index = index vector of B1 samples that should be used % for correction %
function [corr_img]=contrast_B1_correction(img,rel_B1map,B1_input,B1_output,SEGMENT_2D,fit_type,B1_input_index) % [corr_img]=contrast_B1_correction(img,rel_B1map,B1_input,B1_output,SEGMENT_2D,fit_type,B1_input_index) % output: B1 corrected images (2D or 3D-stack) (x,y,B1) % input: img = 3D-stack of images (x,y,B1) which are used for B1-correciton % rel_B1map = relative B1map % B1_output = output B1 (scalar or vector) % SEGMENT_2D = pixel mask for evaluation % fit_type = fit or interpolation type % B1_input_index = index vector of B1 samples that should be used % for correction
b1-correction.txt · Last modified: 2021/10/11 13:22 (external edit)

